Projects I have worked on produced multiple tools that I hope are useful for the science community.
Kosciessa, J. Q., Grandy, T. H., Garrett, D. D., & Werkle-Bergner, M. (2020). Single-trial characterization of neural rhythms: Potential and challenges. NeuroImage, 206, 116331. doi:10.1016/j.neuroimage.2019.116331
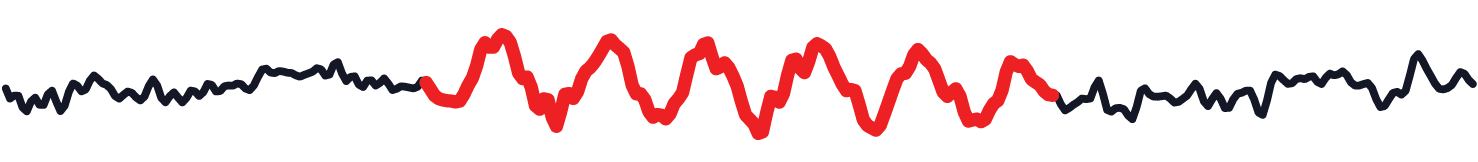
Rhythmic patterns in neural time series provide a window on the neural dynamics that shape perception, cognition and action, and therefore are a major signal of interest in the cognitive, computational and systems neurosciences. However, typical descriptions of rhythmicity lack detail, e.g., failing to indicate when and for how long rhythms occur. Such lack of detail becomes problematic in the face of non-stationarities in rhythmic engagement, i.e., when the presence of rhythms varies over time. To overcome resulting pitfalls, there is a need for methods that identify the occurence of non-stationary rhythmic periods in a systematic manner.
eBOSC (extended Better OSCillation detection) is a toolbox (or a set of scripts) that can be used to detect the occurrence of rhythms in continuous signals (i.e., at the single trial level). It uses a static aperiodic ‘background’ spectrum as the basis to define a ‘power threshold’ that continuous signals have to exceed in order to qualify as ‘rhythmic’. As such, it leverages the observation that stochastic components of the frequency spectrum of neural data are characterized by a ‘1/f’-like power spectrum. An additional ‘duration threshold’ can be set up in advance, or rhythmic episodes can be filtered by duration following detection to ensure that detected rhythmic episodes have a rather sustained vs. transient appearance. The main goal of the code is to produce a list of rhythmic episodes.
The MATLAB code is provided here.
A wiki is provided here.
A python version is available provided on GitHub and on pypi.
Chetverikov, A.+, Kosciessa, J. Q.+, Cornelissen, M., van der Zee, K., & Verhagen, L. (2024). PRESTUS (0.3.0). Zenodo. https://doi.org/10.5281/zenodo.15095861

PRESTUS (PREprocessing & Simulations for Transcranial Ultrasound Stimulation) is an open-source MATLAB toolbox that aims to streamline imaging-informed simulations of Transcanial Ultrasound Stimulation (TUS): from the segmentation of T1-weighted MRI head scans and mapping of medium tissue properties (possibly informed for skull via (pseudo-)CT images) in a simulation grid, to the execution of acoustic and thermal simulations using the widely adopted k-Wave engine. High-performance computing (HPC) support via SLURM and CentOS enables efficient parallelization, and large-scale analyses. Output 3D NifTI images are automatically mapped to a standard template (MNI) space to facilitate group reporting.
The MATLAB code is provided here.
An overview poster is provided here.
An online lecture on personalized simulations is provided here.
Kosciessa, J. Q., Kloosterman, N. A., & Garrett, D. D. (2020). Standard multiscale entropy reflects neural dynamics at mismatched temporal scales: What’s signal irregularity got to do with it? PLoS Computational Biology, 16(5), e1007885. doi:10.1371/journal.pcbi.1007885
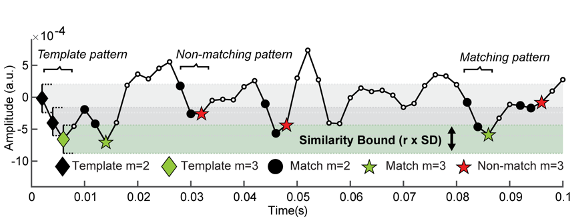
The code is provided here.
A short tutorial is provided here.
Kosciessa, J. Q., Lindenberger, U. & Garrett, D. D. (2020). Thalamocortical excitability adjustments guide human perception under uncertainty. Manuscript submitted for publication.
Multi-attribute-task used in Kosciessa, J. Q., Lindenberger, U., & Garrett, D. D. (2020). Thalamocortical excitability adjustments guide human perception under uncertainty.
The code is provided here.

Voice is a naturalistic response mode in psychological experiments, but labeling thousands of word responses, sometimes from continuous recordings, requires time and patience. CARL has been designed to make the manual labeling process smoother and more time-efficient, as well as to provide measures of voice on- and offset times. It facilitates labeling by (a) splitting long recordings into word chunks (if not already provided by the user as separate words), (b) automatically pre-labeling word on- and offsets based on sound energy, (c) allowing quick manual refinement of sound on- and offsets via drag and drop, (d) providing a few options for label speed-up, e.g., auto-starting only the pre-labeled portions of the recording, (e) allows to manually split chunks containing multiple words into separate segments.
The code is provided here.